BLAST
The BLAST function enables users to input nucleic acid or protein sequences for analysis. It supports the selection of established nucleic acid or protein libraries of Sapindaceae species for alignment, including CDS library, genome library, and protein library. Additionally, the SwissProt library has been added to the protein library to support gene function annotation of any sequence. The BLAST function will automatically determine whether it is a nucleic acid sequence or a protein sequence based on the user's input, and automatically select a matching BLAST mode.
Input file format
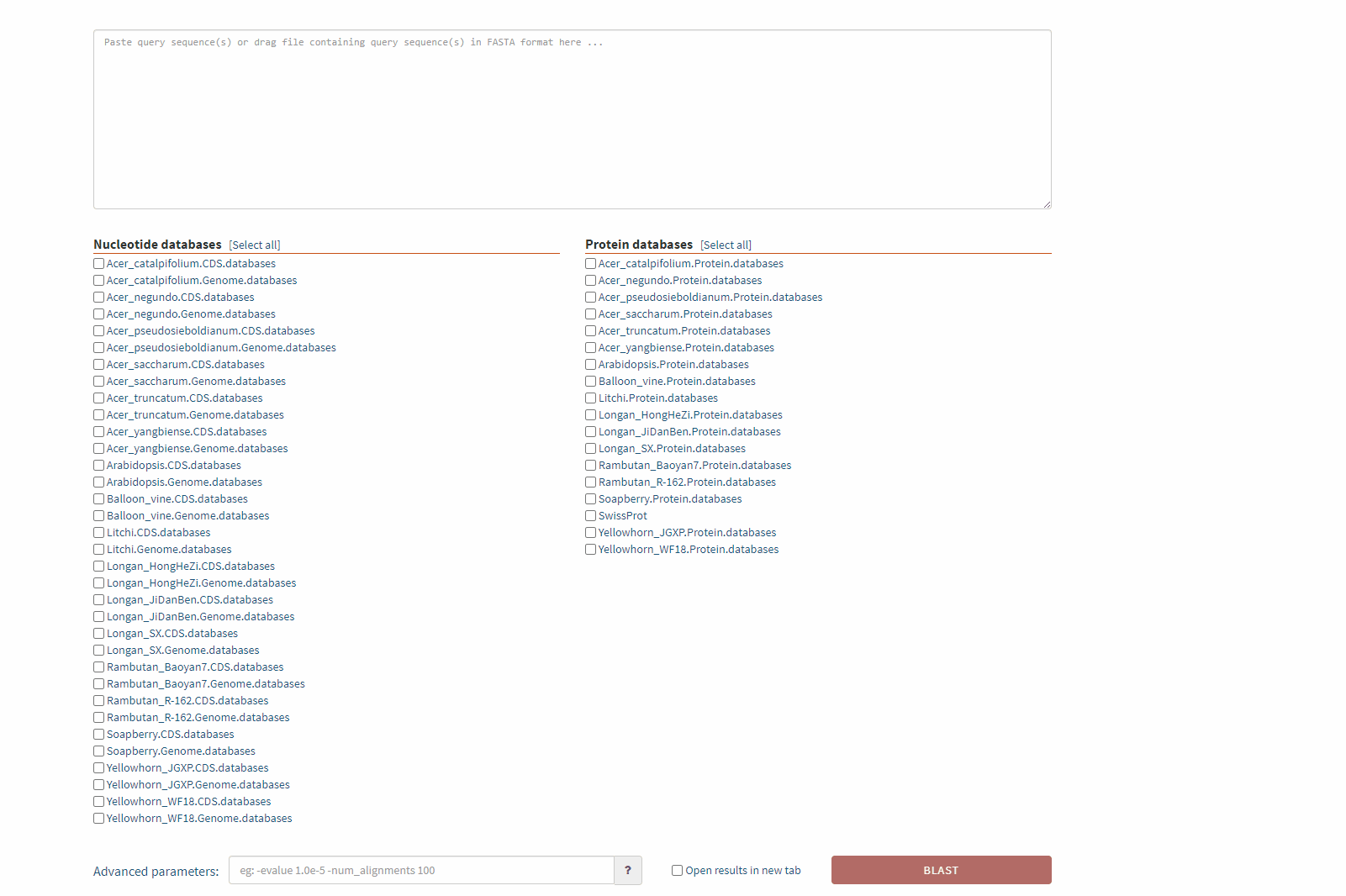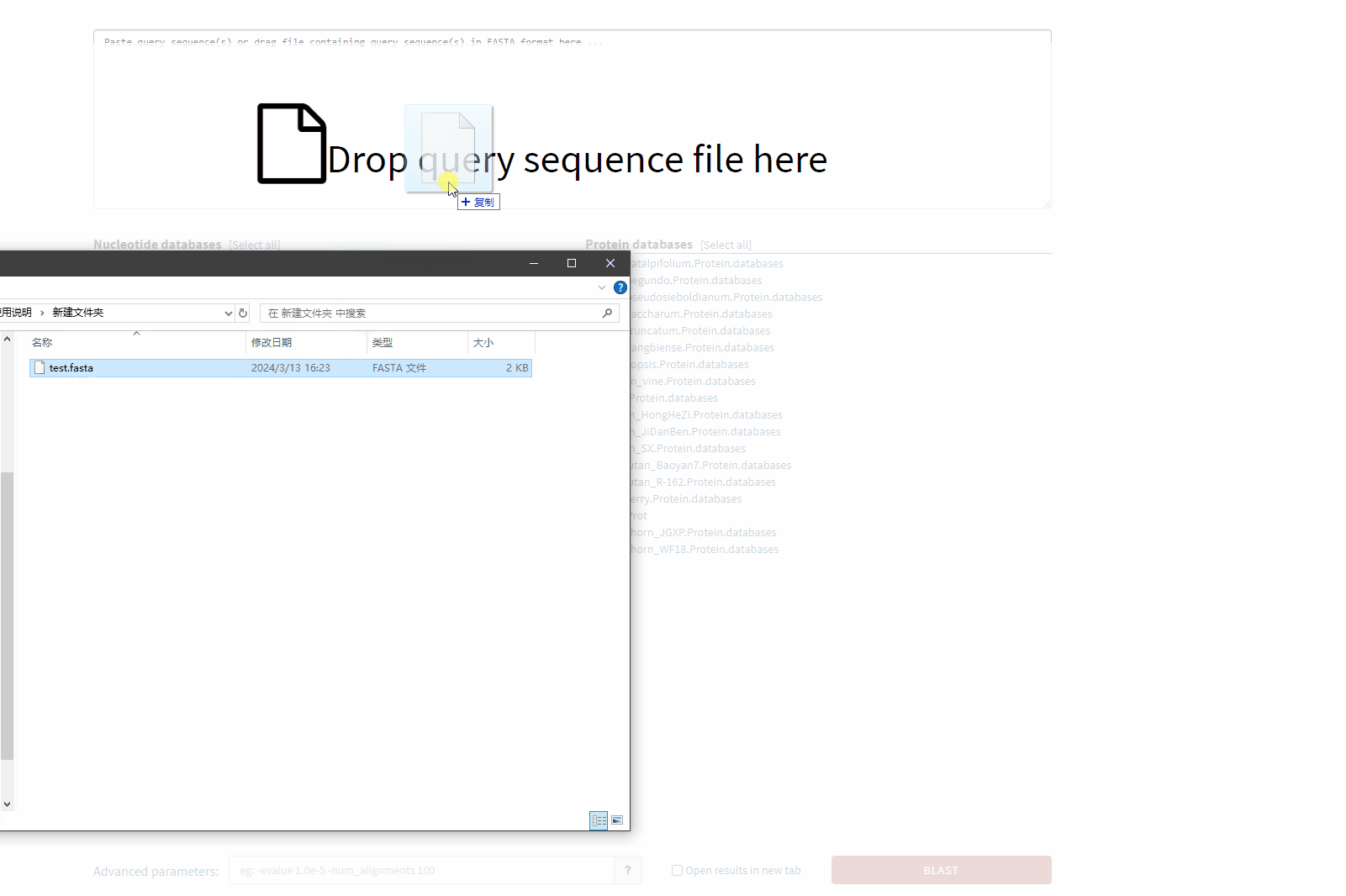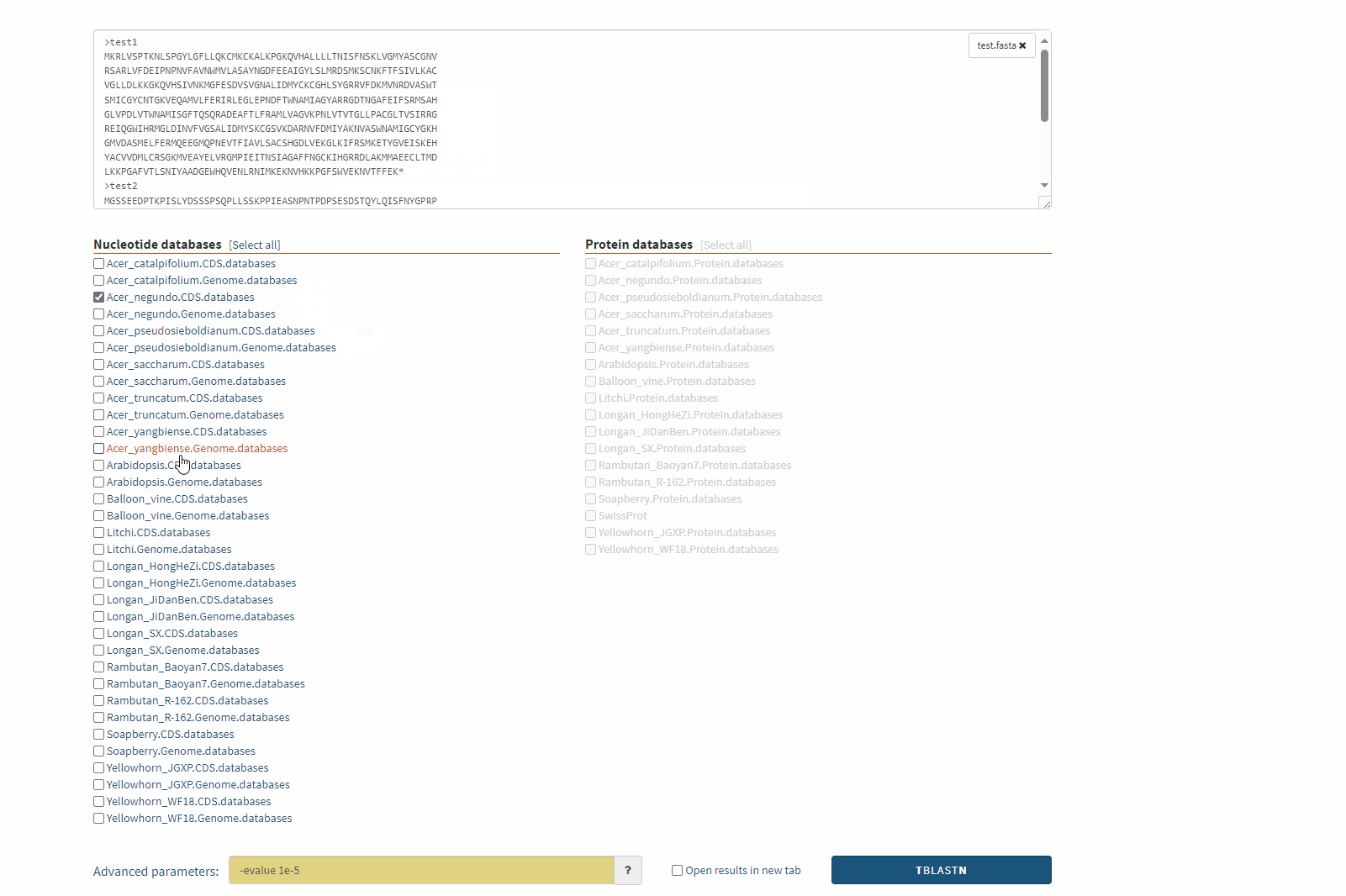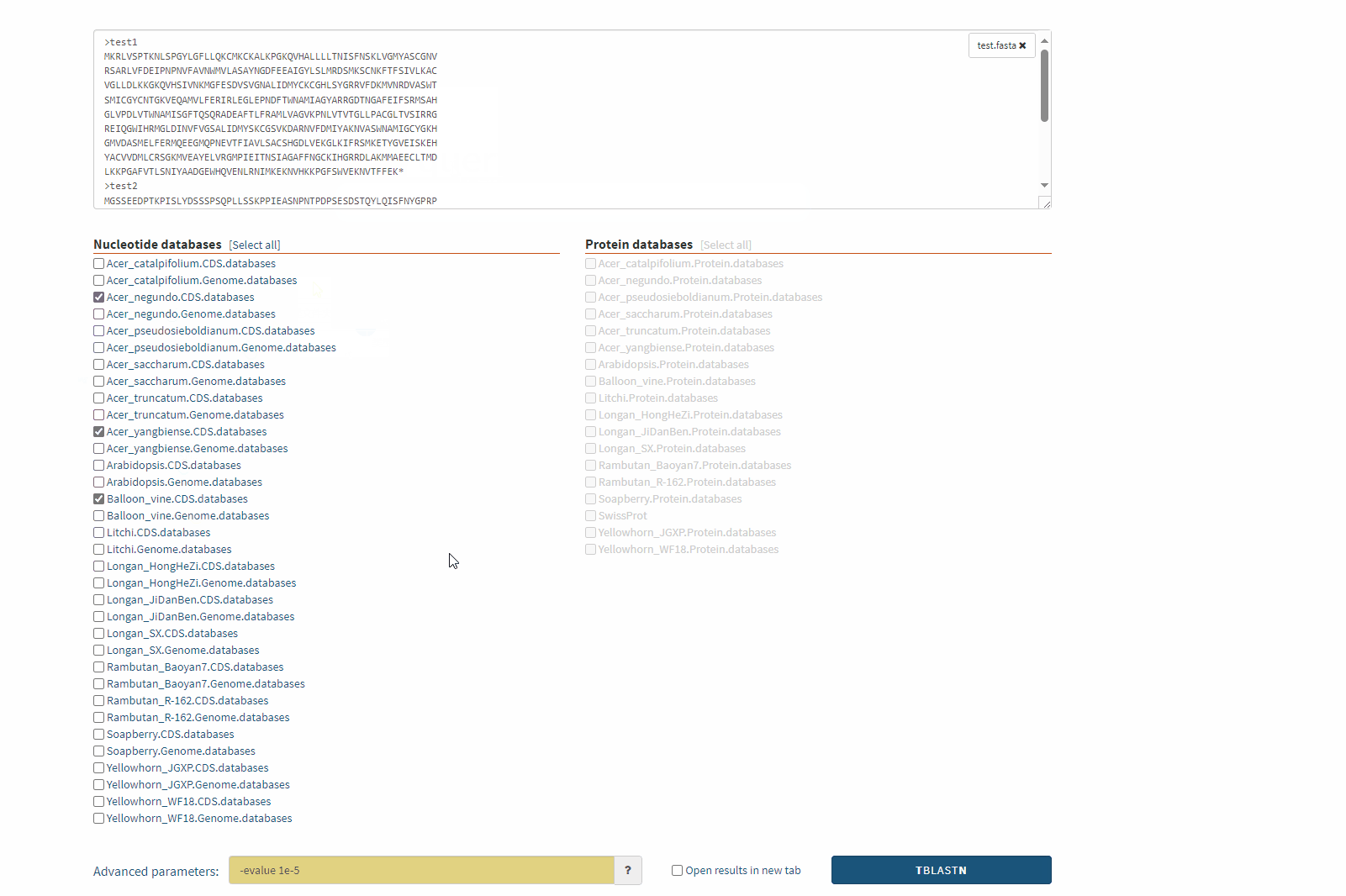
Input a single nucleic acid or protein sequence in the input box. Multiple sequence input (fasta format) is supported. It also supports users to upload data by dragging and dropping files.
Example of single sequence input:
xxxxxxxxxxATGAAAAGACTAGTCTCTCCAACAAAAAACTTATCTCCAGGCTACCTGGGTTTCTTGCTTCAAAAATGCATGAAATGCAAGGCCTTGAAACCAGGAAAGCAAGTTCATGCTTTGTTATTGTTGACCAATATTTCTTTCAACTCCAAGCTTGTTGGCATGTACGCGAGTTGTGGAAATGTCAGGTCTGCGCGCTTGGTGTTCGATGAAATTCCCAACCCAAACGTTTTCGCTGTGAATTGGATGGTTTTGGCCTCCGCTTATAATGGGGACTTCGAAGAGGCAATTGGGTACTTATCGTTGATGCGAGACTCGATGAAATCATGCAACAAGTTTACGTTTTCGATCGTGTTAAAAGCTTGCGTTGGTTTGCTTGATTTGAAGAAAGGAAAACAAGTTCATTCAATTGTGAACAAAATGGGTTTTGAGAGTGATGTCTCAGTTGGTAATGCTTTGATTGATATGTATTGTAAATGCGGACATTTGAGTTATGGTCGTAGAGTGTTTGATAAAATGGTTAACAGAGATGTTGCTTCTTGGACTTCAATGATTTGTGGGTATTGTAACACAGGAAAGGTTGAGCAAGCGATGGTTTTGTTTGAAAGGATAAGGTTGGAAGGCTTAGAACCTAACGATTTCACGTGGAATGCGATGATAGCTGGCTATGCTAGGAGAGGTGATACCAATGGAGCATTTGAGATTTTCTCTAGGATGTCTGCACATGGTCTGGTTCCTGATTTGGTCACTTGGAATGCCATGATTTCAGGTTTCACTCAAAGCCAGCGAGCTGATGAAGCTTTTACATTGTTTCGGGCAATGTTAGTTGCTGGGGTCAAACCGAATCTTGTAACTGTCACGGGACTGCTTCCGGCTTGTGGGCTGACAGTTTCGATTCGAAGAGGTAGAGAAATTCAGGGATGGATACATAGGATGGGGCTTGATATTAATGTATTTGTTGGTAGTGCTCTTATTGACATGTATTCAAAATGTGGGAGTGTGAAAGATGCTAGGAATGTGTTTGAC
or
xxxxxxxxxxMKRLVSPTKNLSPGYLGFLLQKCMKCKALKPGKQVHALLLLTNISFNSKLVGMYASCGNVRSARLVFDEIPNPNVFAVNWMVLASAYNGDFEEAIGYLSLMRDSMKSCNKFTFSIVLKACVGLLDLKKGKQVHSIVNKMGFESDVSVGNALIDMYCKCGHLSYGRRVFDKMVNRDVASWTSMICGYCNTGKVEQAMVLFERIRLEGLEPNDFTWNAMIAGYARRGDTNGAFEIFSRMSAHGLVPDLVTWNAMISGFTQSQRADEAFTLFRAMLVAGVKPNLVTVTGLLPACGLTVSIRRGREIQGWIHRMGLDINVFVGSALIDMYSKCGSVKDARNVFDMIYAKNVASWNAMIGCYGKHGMVDASMELFERMQEEGMQPNEVTFIAVLSACSHGDLVEKGLKIFRSMKETYGVEISKEHYACVVDMLCRSGKMVEAYELVRGMPIEITNSIAGAFFNGCKIHGRRDLAKMMAEECLTMDLKKPGAFVTLSNIYAADGEWHQVENLRNIMKEKNVHKKPGFSWVEKNVTFFEK*
Example of multiple sequences input:
xxxxxxxxxx>test1MKRLVSPTKNLSPGYLGFLLQKCMKCKALKPGKQVHALLLLTNISFNSKLVGMYASCGNVRSARLVFDEIPNPNVFAVNWMVLASAYNGDFEEAIGYLSLMRDSMKSCNKFTFSIVLKACVGLLDLKKGKQVHSIVNKMGFESDVSVGNALIDMYCKCGHLSYGRRVFDKMVNRDVASWTSMICGYCNTGKVEQAMVLFERIRLEGLEPNDFTWNAMIAGYARRGDTNGAFEIFSRMSAHGLVPDLVTWNAMISGFTQSQRADEAFTLFRAMLVAGVKPNLVTVTGLLPACGLTVSIRRGREIQGWIHRMGLDINVFVGSALIDMYSKCGSVKDARNVFDMIYAKNVASWNAMIGCYGKHGMVDASMELFERMQEEGMQPNEVTFIAVLSACSHGDLVEKGLKIFRSMKETYGVEISKEHYACVVDMLCRSGKMVEAYELVRGMPIEITNSIAGAFFNGCKIHGRRDLAKMMAEECLTMDLKKPGAFVTLSNIYAADGEWHQVENLRNIMKEKNVHKKPGFSWVEKNVTFFEK*>test2MGSSEEDPTKPISLYDSSSPSQPLLSSKPPIEASNPNTPDPSESDSTQYLQISFNYGPRPFKDLPFLLLFLLFVLSTFGFGIFAICNRNSNYTNVSAFVYSSTSGSCVKNSTSPSFSLFSSFVHVGSGFSLSKSHLLRDLIWTLVMTGVLSVPICFLLLLLLKRYTKQIVYICLPFFIIIPVFINVYWFVACSISSTCGDAFPLAYRILVFVFAFLIIGVIVWILVVNWHRIELTVMIIGVASDALSKNLGLFAVLPLLTLGLVVYYAPIVVFLVFVRFNGKIIPKESNGEYTCVWKQDSWVPAYFTLAILTMLWSLTVMVEAQAYVISGTTAQWYFSKEDKKPRRSIRSSLRNAFGPSSGTVCLSGLLICVVRIVRAAVDSARQEDVPGFVNLVLRCCVNALLSAIDFLNKFTINFAAITGEAYCSSARMTYEVLKRNLLSAVFVETVSTRLLGGIIFVLSALYAIAVCAILQGATKLGVDSYFVAVLAWVLLLVVLAYFVHVLDNVIDTVYICYAIDRDRGEVYKADVHEVYVHLPISRNHAASYAPRTVGA.
Example of drag-and-drop file upload sequence:
Output file format
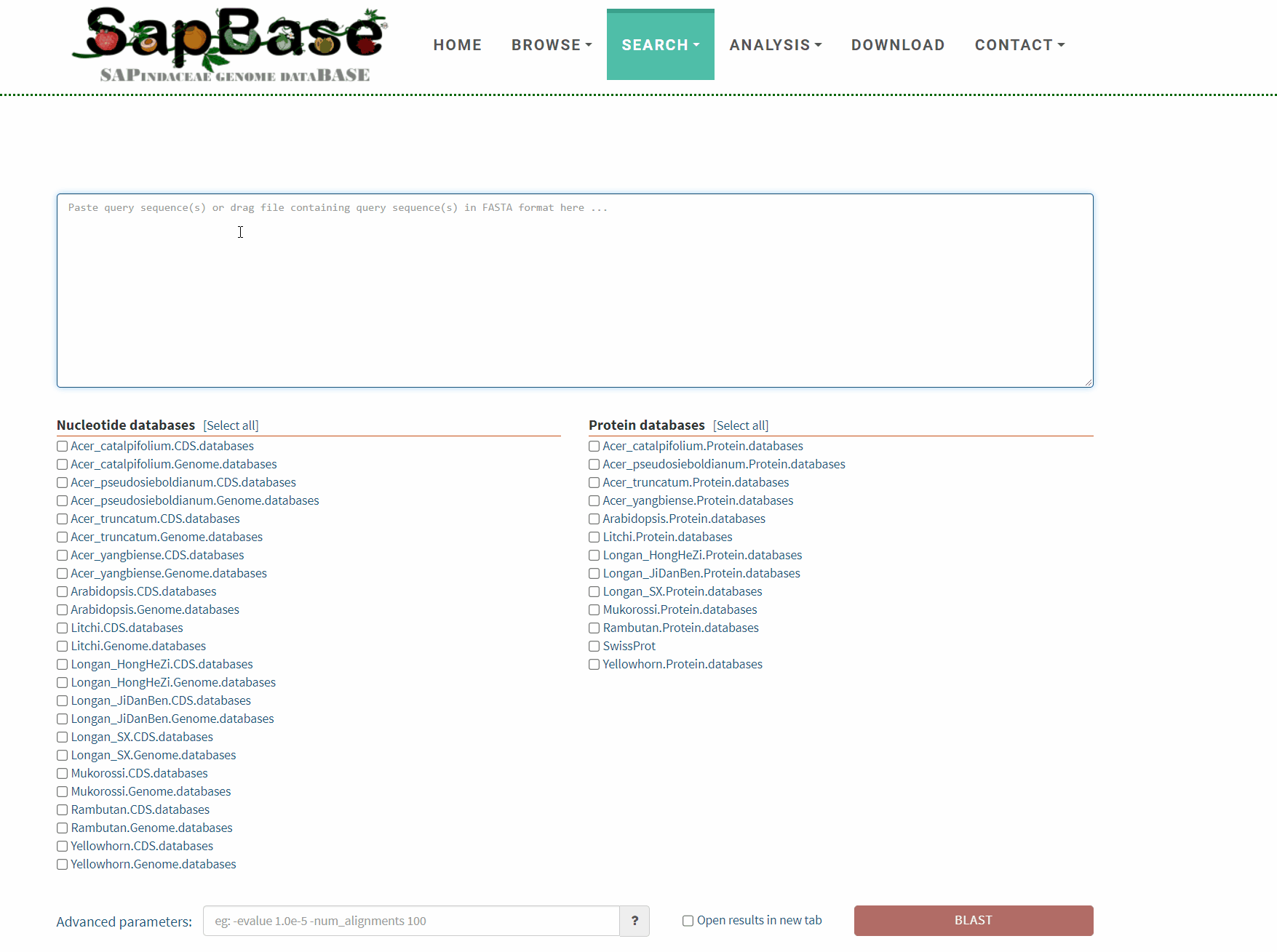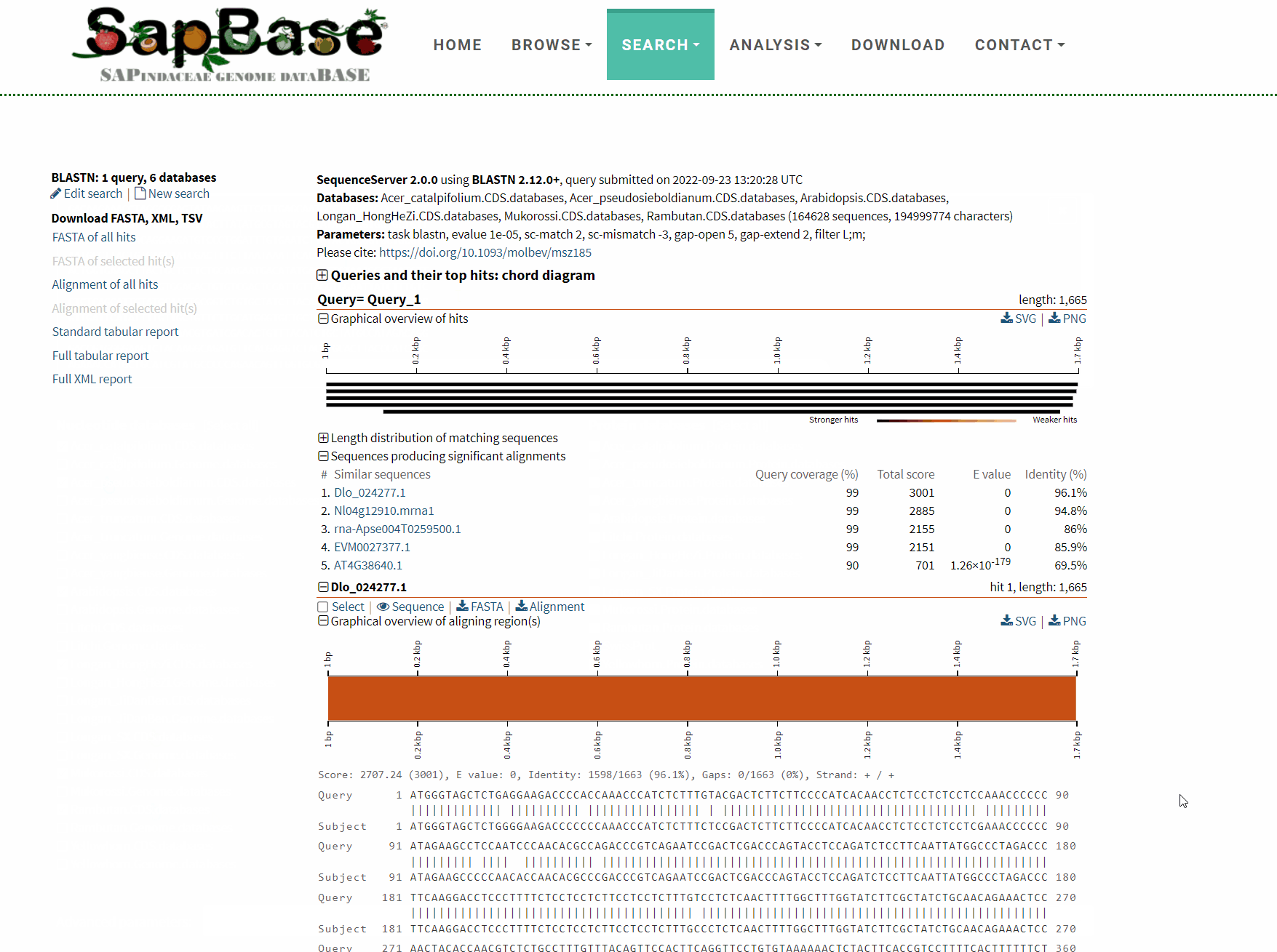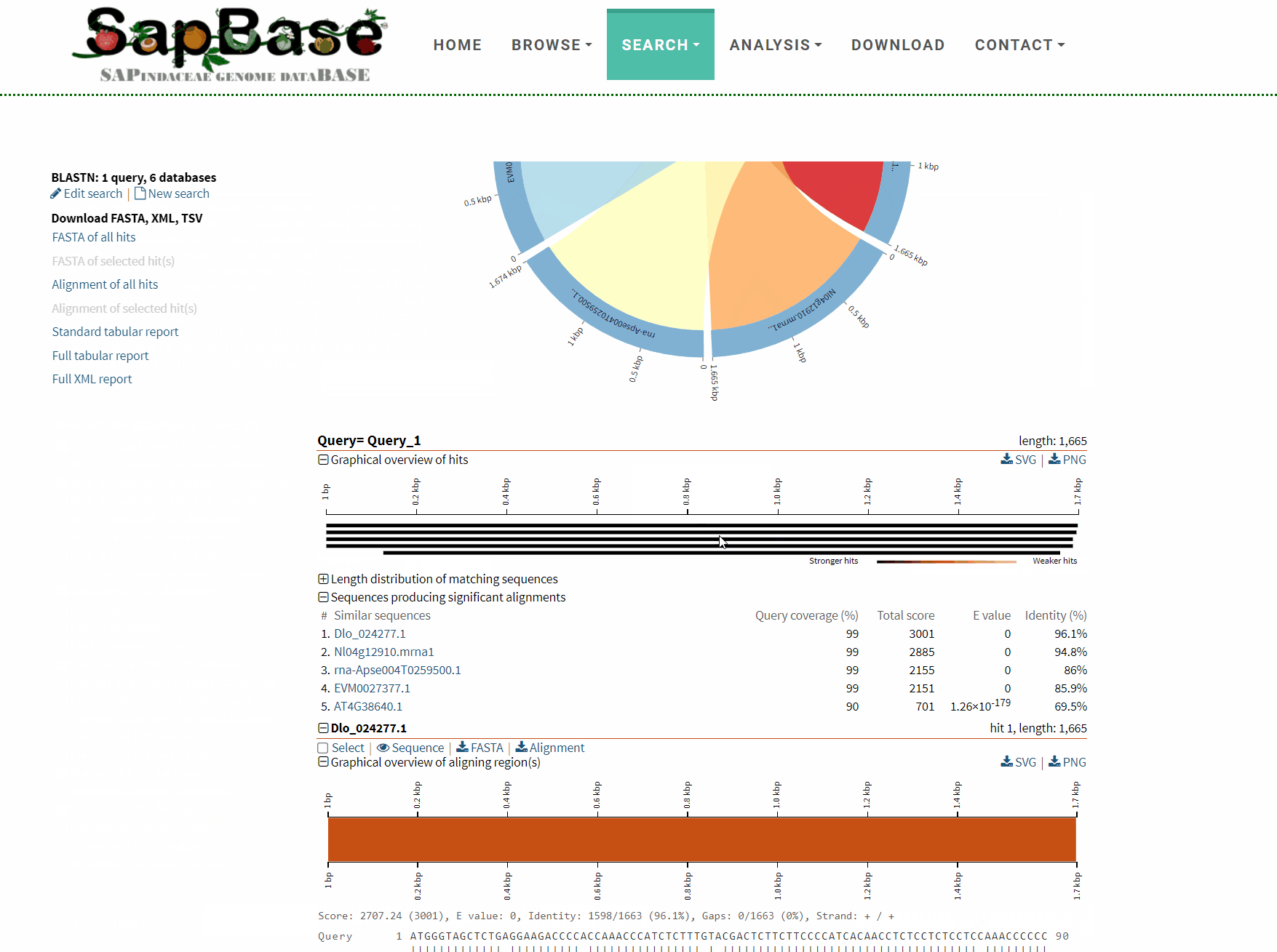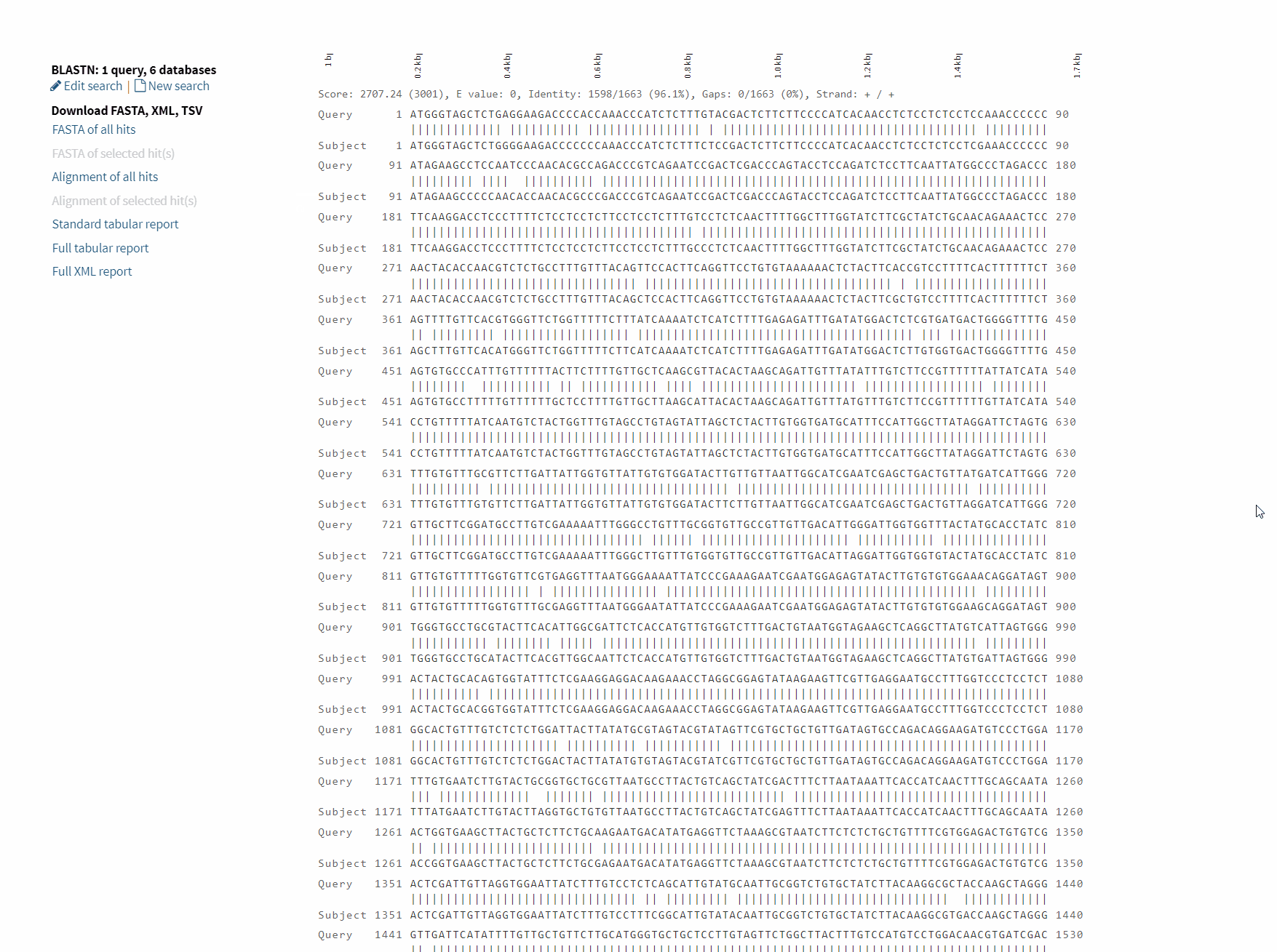
After BLAST is completed, an interactive results page will be returned, and users can choose to download or visualize the corresponding results as needed.

Detailed usage demonstration of BLAST function: