The chromosome-level rambutan genome reveals a significant role of segmental duplication in the expansion of resistance genes
Link to paper: https://doi.org/10.1093/hr/uhac014
Published: 11 April 2022, Horticulture Research
ABSTRACT
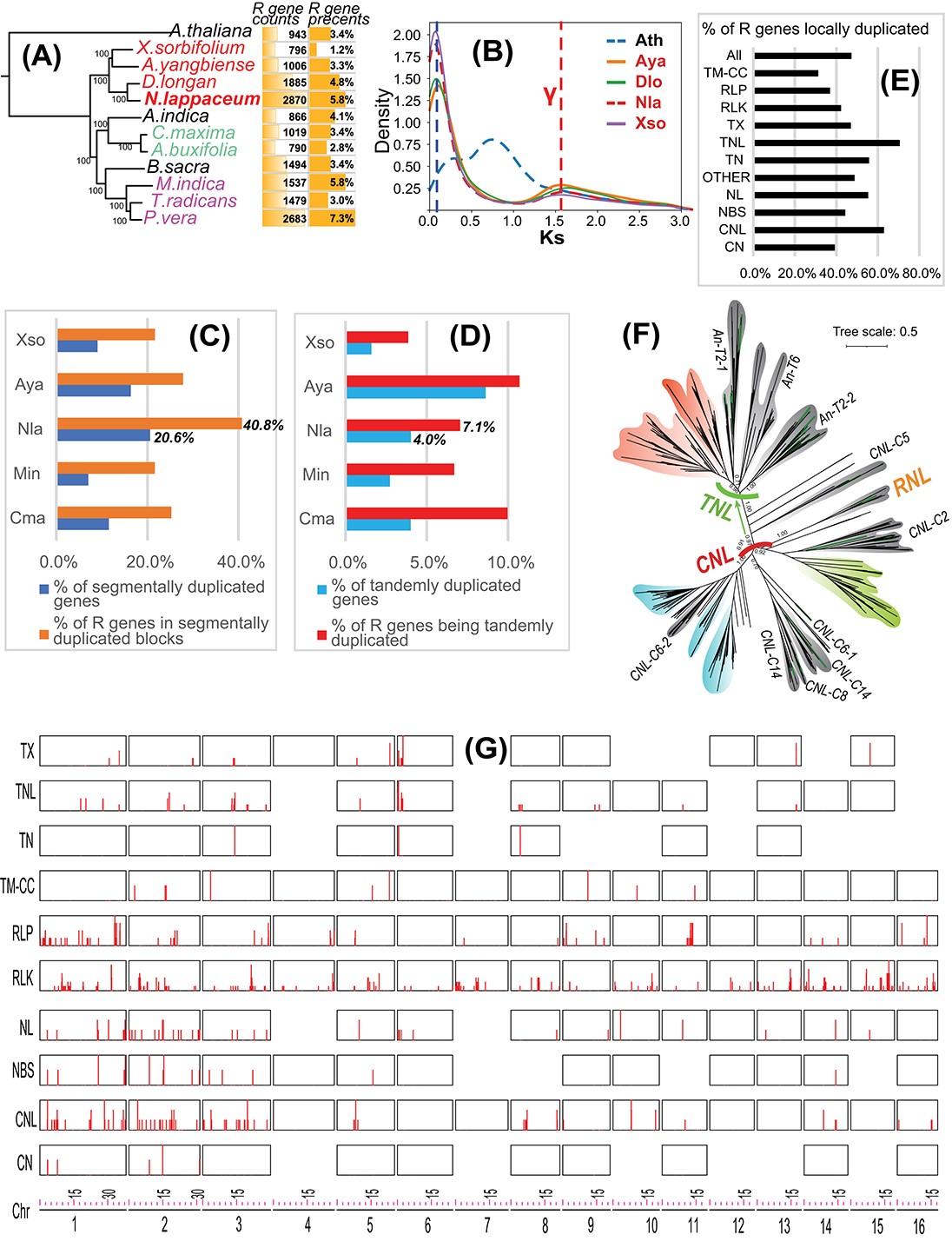
Rambutan (Nephelium lappaceum var. lappaceum), a tropical fruit tree native to southeastern Asia, belongs to the family Sapindaceae. Rambutan is a popular table fruit and is also processed into preserves, juices, wines, and sorbets. At present, only three Sapindaceae genomes are publicly available: Xanthoceras sorbifolium, Dimocarpus longan (longan), and Acer yangbiense. During the process of submitting this manuscript, the genome paper for the rambutan cultivar Baoyan7 became available online, but its genome sequence has not yet been released.
Here, we sequenced the rambutan cultivar R-162 sampled from Puerto Rico (USDA-ARS, Tropical Agriculture Research Station, Mayaguez) in order to study the genomic expansion of plant resistance genes. The R-162 genome has a scaffold N50 of 21.65 Mb and 16 pseudo-chromosomes. The chromosome-level genome was assembled using Dovetail Genomics HiRise scaffolding software with long-range Chicago libraries and Hi-C library sequencing. Genome annotation identified 49,959 protein-coding genes, 38,075 of which were annotated by eggNOG and 33,429 of which were supported by RNA-seq data. BUSCO (Benchmarking Universal Single-Copy Orthologs) analysis identified 1,573 single-copy orthologous genes, corresponding to a genome completeness of 97.5%. The genome and annotation data can be accessed at https://bcb.unl.edu/Nla/ and NCBI (BioProject ID: PRJNA766632) (Zheng et al., 2022).