De novo genome assembly of the endangered Acer yangbiense, a plant species with extremely small populations endemic to Yunnan Province, China
Link to paper: https://doi.org/10.1093/gigascience/giz085
Published: 15 July 2019, GigaScience
ABSTRACT
Background
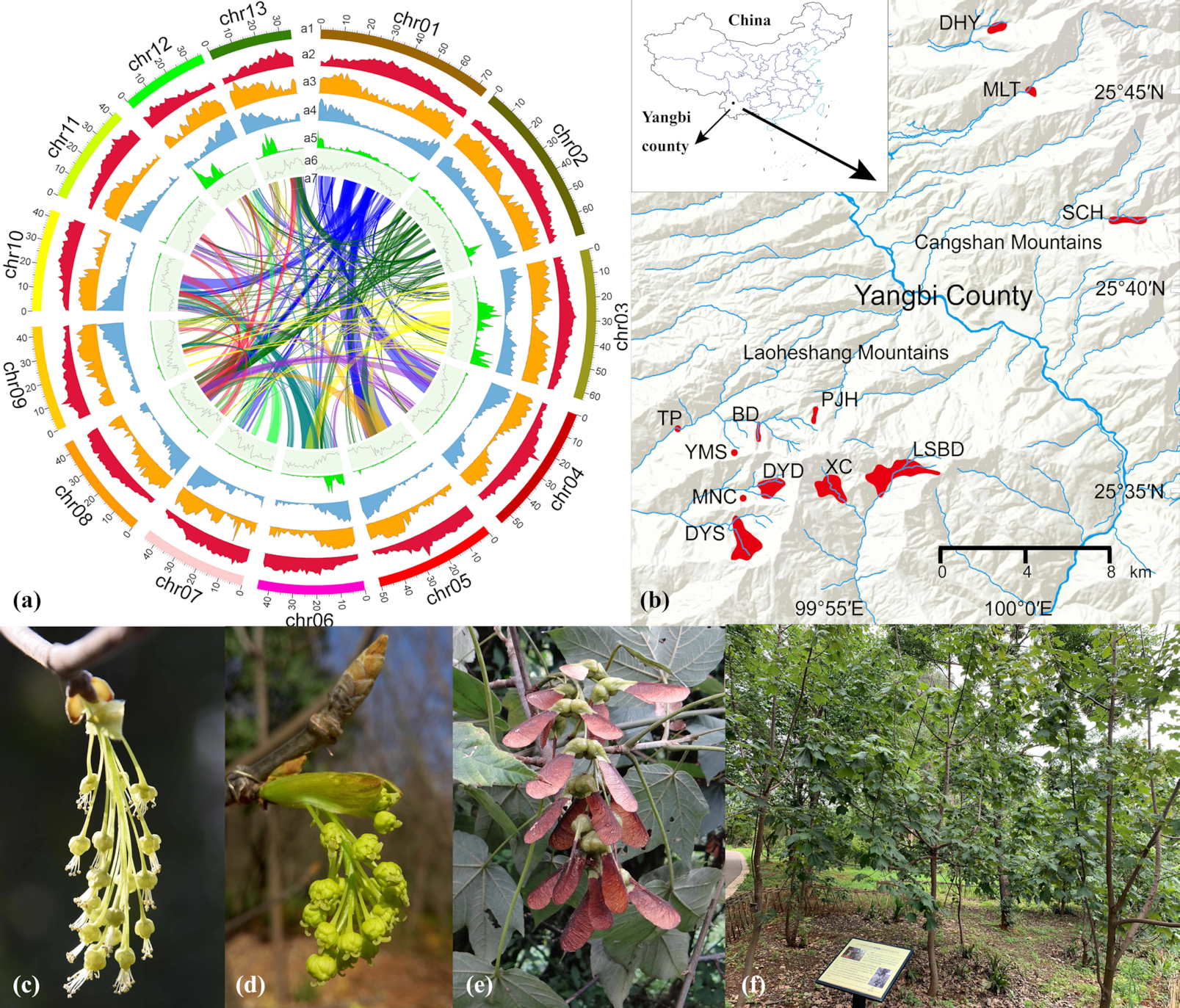
Acer yangbiense is a newly described critically endangered endemic maple tree confined to Yangbi County in Yunnan Province in Southwest China. It was included in a programme for rescuing the most threatened species in China, focusing on “plant species with extremely small populations (PSESP)”.
Findings
We generated 64, 94, and 110 Gb of raw DNA sequences and obtained a chromosome-level genome assembly of A. yangbiense through a combination of Pacific Biosciences Single-molecule Real-time, Illumina HiSeq X, and Hi-C mapping, respectively. The final genome assembly is ∼666 Mb, with 13 chromosomes covering ∼97% of the genome and scaffold N50 sizes of 45 Mb. Further, BUSCO analysis recovered 95.5% complete BUSCO genes. The total number of repetitive elements account for 68.0% of the A. yangbiense genome. Genome annotation generated 28,320 protein-coding genes, assisted by a combination of prediction and transcriptome sequencing. In addition, a nearly 1:1 orthology ratio of dot plots of longer syntenic blocks revealed a similar evolutionary history between A. yangbiense and grape, indicating that the genome has not undergone a whole-genome duplication event after the core eudicot common hexaploidization.
Conclusion
Here, we report a high-quality de novo genome assembly of A. yangbiense, the first genome for the genus Acer and the family Aceraceae. This will provide fundamental conservation genomics resources, as well as representing a new high-quality reference genome for the economically important Acer lineage and the wider order of Sapindales (Yang et al., 2019).